XArray Example#
To run this example, an environment.yml similar to this one could be used:
name: bh_xhistogram
channels:
- conda-forge
dependencies:
- python==3.8
- boost-histogram
- xhistogram
- matplotlib
- netcdf4
[1]:
import matplotlib.pyplot as plt
import numpy as np
import xarray as xr
from xhistogram.xarray import histogram as xhistogram
import boost_histogram as bh
Let’s look at using boost-histogram to imitate the xhistogram package by reading and producing xarrays. As a reminder, xarray is a sort of generalized Pandas library, supporting ND labeled and indexed data.
Simple 1D example#
We will start with the first example from the xhistogram docs:
[2]:
da = xr.DataArray(np.random.randn(100, 30), dims=["time", "x"], name="foo")
bins = np.linspace(-4, 4, 20)
xhistogram#
And, this is what historamming and plotting looks like:
[3]:
h = xhistogram(da, bins=[bins])
display(h)
h.plot()
# h is an xarray
<xarray.DataArray 'histogram_foo' (foo_bin: 19)>
array([ 2, 1, 2, 22, 56, 135, 234, 375, 453, 496, 456, 346, 226,
96, 65, 25, 7, 3, 0])
Coordinates:
* foo_bin (foo_bin) float64 -3.789 -3.368 -2.947 -2.526 ... 2.947 3.368 3.789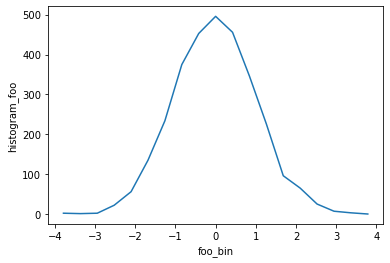
boost-histogram (direct usage)#
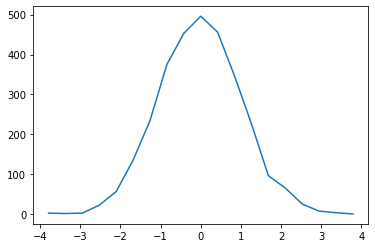
Let’s first just try this by hand, to see how it works. This will not return an xarray, etc.
[4]:
bh_bins = bh.axis.Regular(19, -4, 4)
bh_hist = bh.Histogram(bh_bins).fill(np.asarray(da).flatten())
plt.plot(bh_hist.axes[0].centers, bh_hist.values());
boost-histogram (adapter function)#
Now, let’s make an adaptor for boost-histogram.
[5]:
def bh_xhistogram(*args, bins):
# Convert bins to boost-histogram axes first
prepare_bins = (bh.axis.Variable(b) for b in bins)
h = bh.Histogram(*prepare_bins)
# We need flat NP arrays for filling
prepare_fill = (np.asarray(a).flatten() for a in args)
h.fill(*prepare_fill)
# Now compute the xarray output.
return xr.DataArray(
h.values(),
name="_".join(a.name for a in args) + "_histogram",
coords=[
(f"{a.name}_bin", arr.flatten(), a.attrs)
for a, arr in zip(args, h.axes.centers)
],
)
[6]:
h = bh_xhistogram(da, bins=[bins])
display(h)
h.plot();
<xarray.DataArray 'foo_histogram' (foo_bin: 19)>
array([ 2., 1., 2., 22., 56., 135., 234., 375., 453., 496., 456.,
346., 226., 96., 65., 25., 7., 3., 0.])
Coordinates:
* foo_bin (foo_bin) float64 -3.789 -3.368 -2.947 -2.526 ... 2.947 3.368 3.789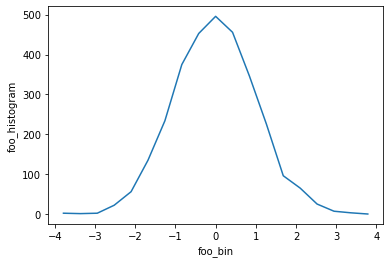
More features#
Let’s add a few more features to our function defined above. * Let’s allow bins to be a list of axes or even a completely prepared histogram; this will allow us to take advantage of boost-histogram features later. * Let’s add a weights keyword so we can do weighted histograms as well.
[7]:
def bh_xhistogram(*args, bins, weights=None):
"""
bins is either a histogram, a list of axes, or a list of bins
"""
if isinstance(bins, bh.Histogram):
h = bins
else:
prepare_bins = (
b if isinstance(b, bh.axis.Axis) else bh.axis.Variable(b) for b in bins
)
h = bh.Histogram(*prepare_bins)
prepare_fill = (np.asarray(a).flatten() for a in args)
if weights is None:
h.fill(*prepare_fill)
else:
prepared_weights, *_ = xr.broadcast(weights, *args)
h.fill(*prepare_fill, weight=np.asarray(prepared_weights).flatten())
return xr.DataArray(
h.values(),
name="_".join(a.name for a in args) + "_histogram",
coords=[
(f"{a.name}_bin", arr.flatten(), a.attrs)
for a, arr in zip(args, h.axes.centers)
],
)
2D example#
This also comes from the xhistogram docs.
[8]:
# Read WOA using opendap
Temp_url = (
"http://apdrc.soest.hawaii.edu:80/dods/public_data/WOA/WOA13/5_deg/annual/temp"
)
Salt_url = (
"http://apdrc.soest.hawaii.edu:80/dods/public_data/WOA/WOA13/5_deg/annual/salt"
)
Oxy_url = (
"http://apdrc.soest.hawaii.edu:80/dods/public_data/WOA/WOA13/5_deg/annual/doxy"
)
ds = xr.merge(
[
xr.open_dataset(Temp_url).tmn.load(),
xr.open_dataset(Salt_url).smn.load(),
xr.open_dataset(Oxy_url).omn.load(),
]
)
[9]:
sbins = np.arange(31, 38, 0.025)
tbins = np.arange(-2, 32, 0.1)
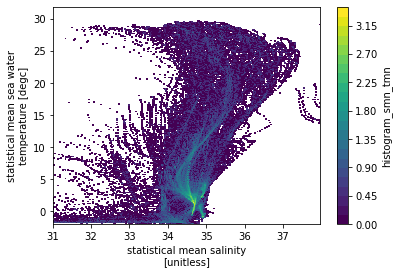
xhistogram#
[10]:
hTS = xhistogram(ds.smn, ds.tmn, bins=[sbins, tbins])
np.log10(hTS.T).plot(levels=31)
/usr/local/Caskroom/miniconda/base/envs/xtest/lib/python3.8/site-packages/xarray/core/computation.py:601: RuntimeWarning: divide by zero encountered in log10
result_data = func(*input_data)
[10]:
<matplotlib.collections.QuadMesh at 0x7f84c920b130>
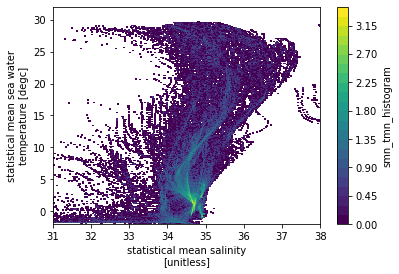
boost-histogram#
We could hand in the same bin definitions, but let’s use boost-histogram axes instead:
[11]:
sax = bh.axis.Regular(250, 31, 38)
tax = bh.axis.Regular(340, -2, 32)
hTS = bh_xhistogram(ds.smn, ds.tmn, bins=[sax, tax])
np.log10(hTS.T).plot(levels=31)
[11]:
<matplotlib.collections.QuadMesh at 0x7f8528c132b0>
Speed comparson#
[12]:
%%timeit
hTS = xhistogram(ds.smn, ds.tmn, bins=[sbins, tbins])
17.6 ms ± 508 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
[13]:
%%timeit
hTS = bh_xhistogram(ds.smn, ds.tmn, bins=[sax, tax])
4.7 ms ± 245 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
Weighted histogram#
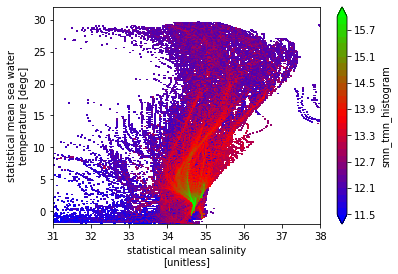
Let’s try a more complex example from the docs; the dVol weights one:
[14]:
dz = np.diff(ds.lev)
dz = np.insert(dz, 0, dz[0])
dz = xr.DataArray(dz, coords={"lev": ds.lev}, dims="lev")
dVol = dz * (5 * 110e3) * (5 * 110e3 * np.cos(ds.lat * np.pi / 180))
hTSw = bh_xhistogram(ds.smn, ds.tmn, bins=[sax, tax], weights=dVol)
np.log10(hTSw.T).plot(levels=31, vmin=11.5, vmax=16, cmap="brg")
[14]:
<matplotlib.collections.QuadMesh at 0x7f84b821c940>